Potential optimizers
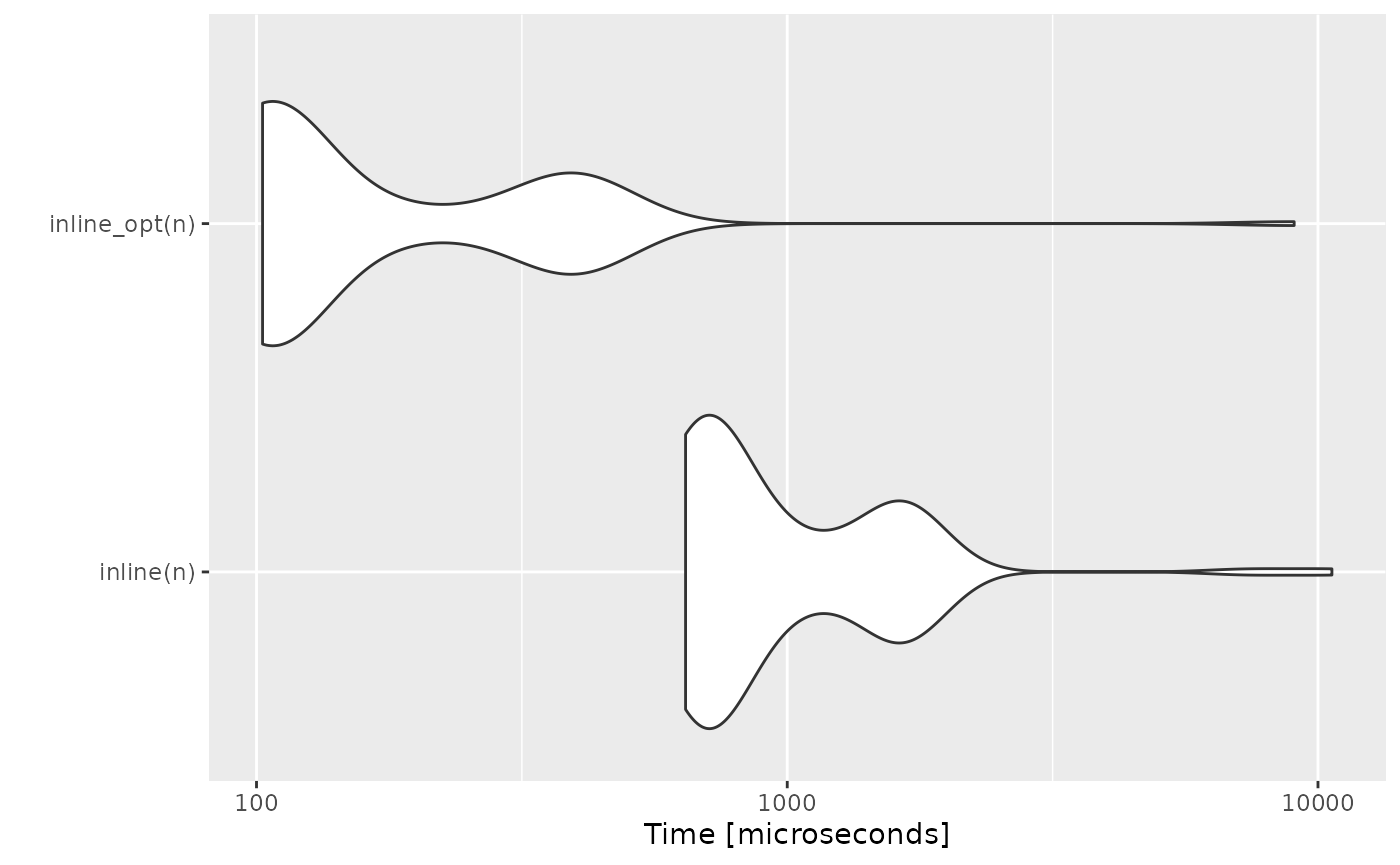
Inline Expansion
Idea
Replacing a function call with the body of the called function is called “inline expansion”. This eliminates the function calling overhead and also the overhead of return call from a function. It also saves the overhead of variables push/pop on the stack while function calling.
Code Examples
Unoptimized Code
cubed <- function(x) {
x * x * x
}
inline <- function(n) {
to_cubes <- 0
for (i in seq_len(n)) {
to_cubes <- to_cubes + cubed(i)
}
}Proposed Optimized Code
inline_opt <- function(n) {
to_cubes <- 0
for (i in seq_len(n)) {
to_cubes <- to_cubes + (i * i * i) # function inlined
}
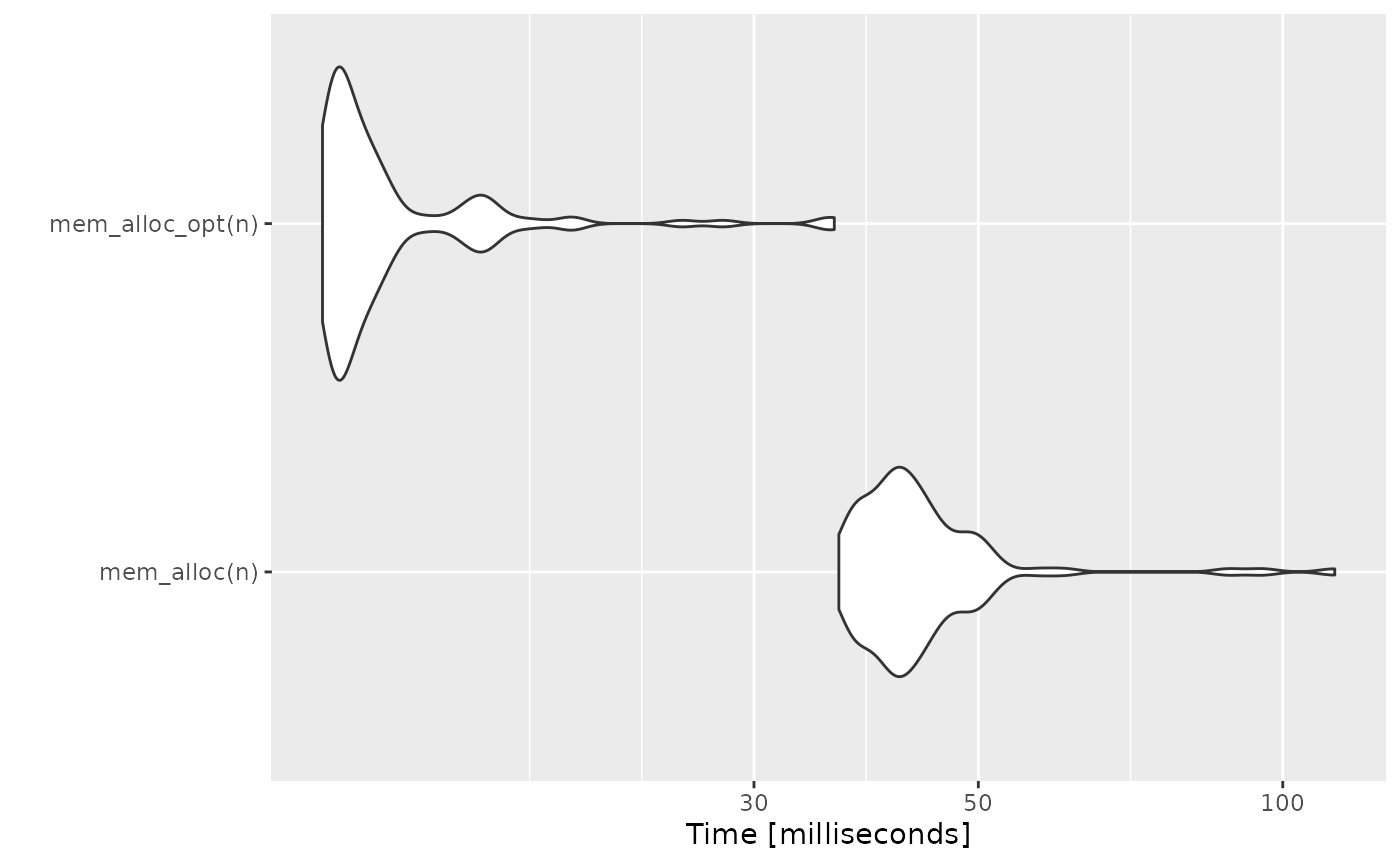
}Memory Pre-Allocation
Idea
As a general rule of thumb, in any programming language, we should undertake memory management as much as possible. When we grow a vector inside a loop, the vector asks the processor for extra space in between the running program and then proceeds, once it gets the required memory. This process is repeated for every iteration of the loop. Thus we should pre-allocate the required memory to a vector to avoid such delays.
Code Examples
Unoptimized Code
mem_alloc <- function(n) {
vec <- NULL
for (i in seq_len(n)) {
vec[i] <- i
}
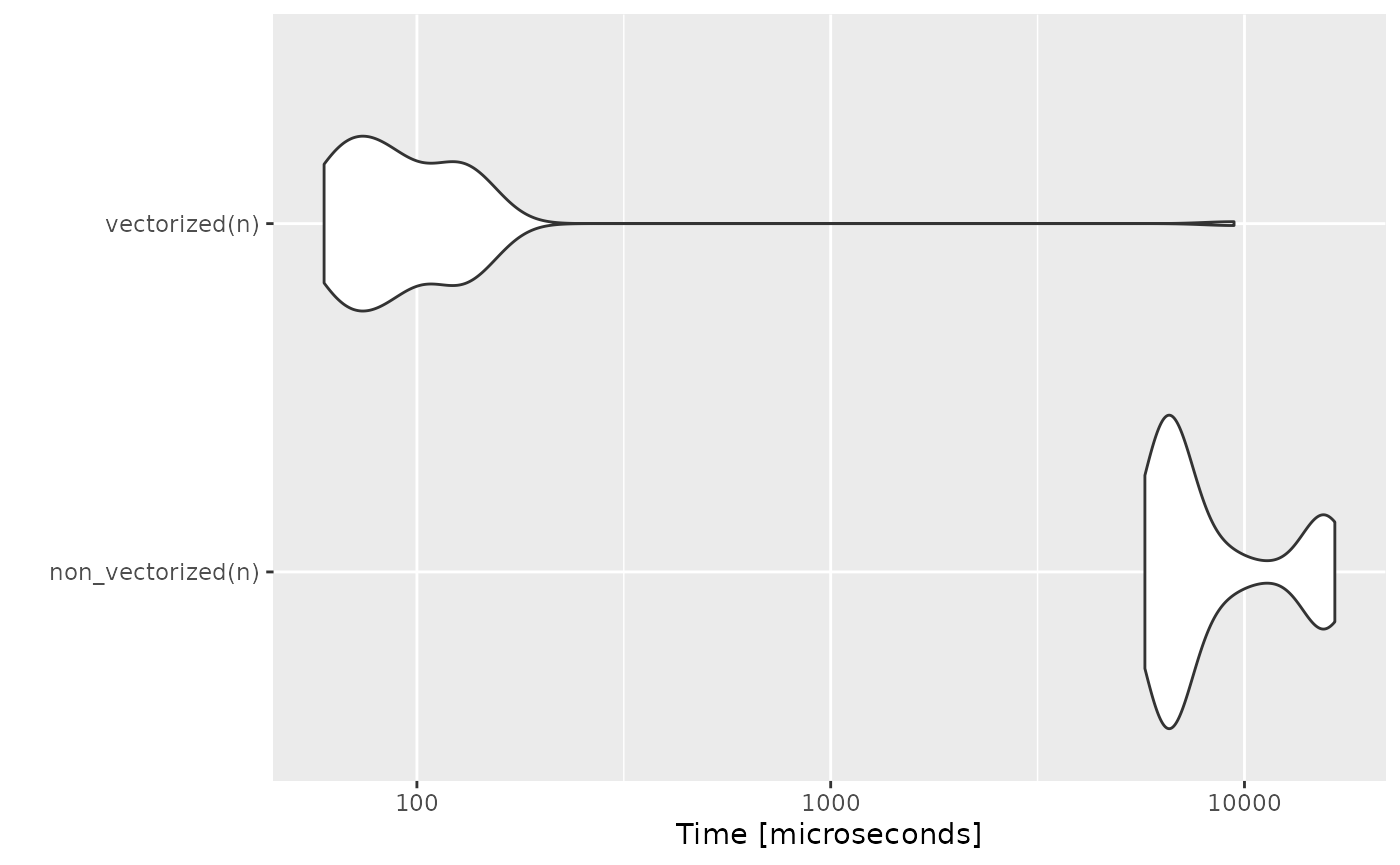
}Vectorization
Idea
A golden rule in R programming is to access the underlying C/Fortran routines as much as possible; the fewer R function calls required to achieve this, the better. Many R functions are therefore vectorized, that is, the function’s inputs and/or outputs naturally work with vectors, reducing the number of function calls required.
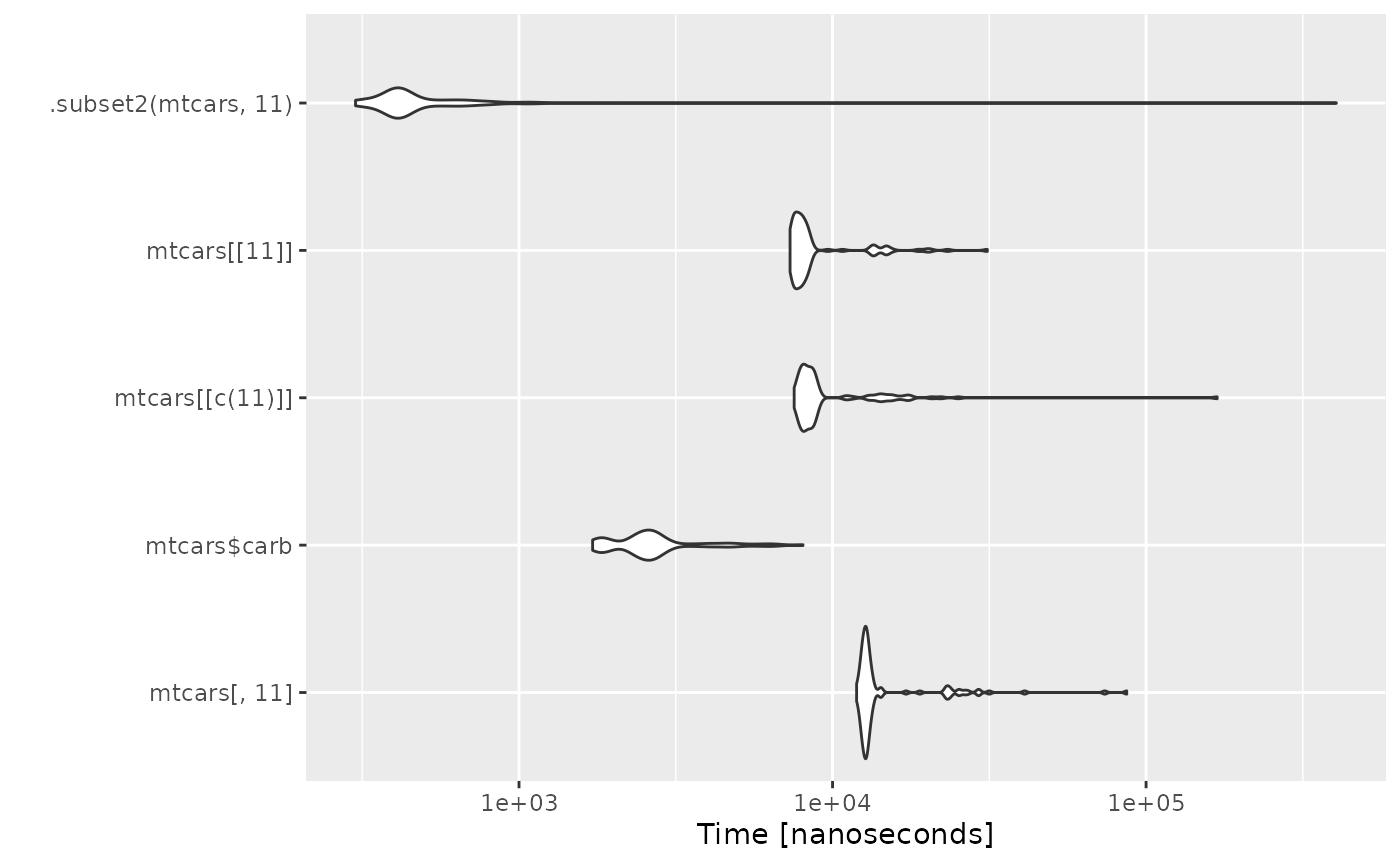
Efficient Column Extraction
Idea
The idea would be to replace the different one-column extraction alternatives by the much faster .subset2 call alternative.
Benchmark
autoplot(microbenchmark(
mtcars[, 11],
mtcars$carb,
mtcars[[c(11)]],
mtcars[[11]],
.subset2(mtcars, 11)
))
Drawbacks
For some R classes, the
[[ ]]operator and.subsetwork differently. For instance, they seem to be equivalent fordata.framebut are not the same formatrixclass.Moreover, both
[[ ]]and.subset2are functions and in R, any function can be overwritten. Thus the above optimization can be made to fail just by redefining, say, the the.subset2function.
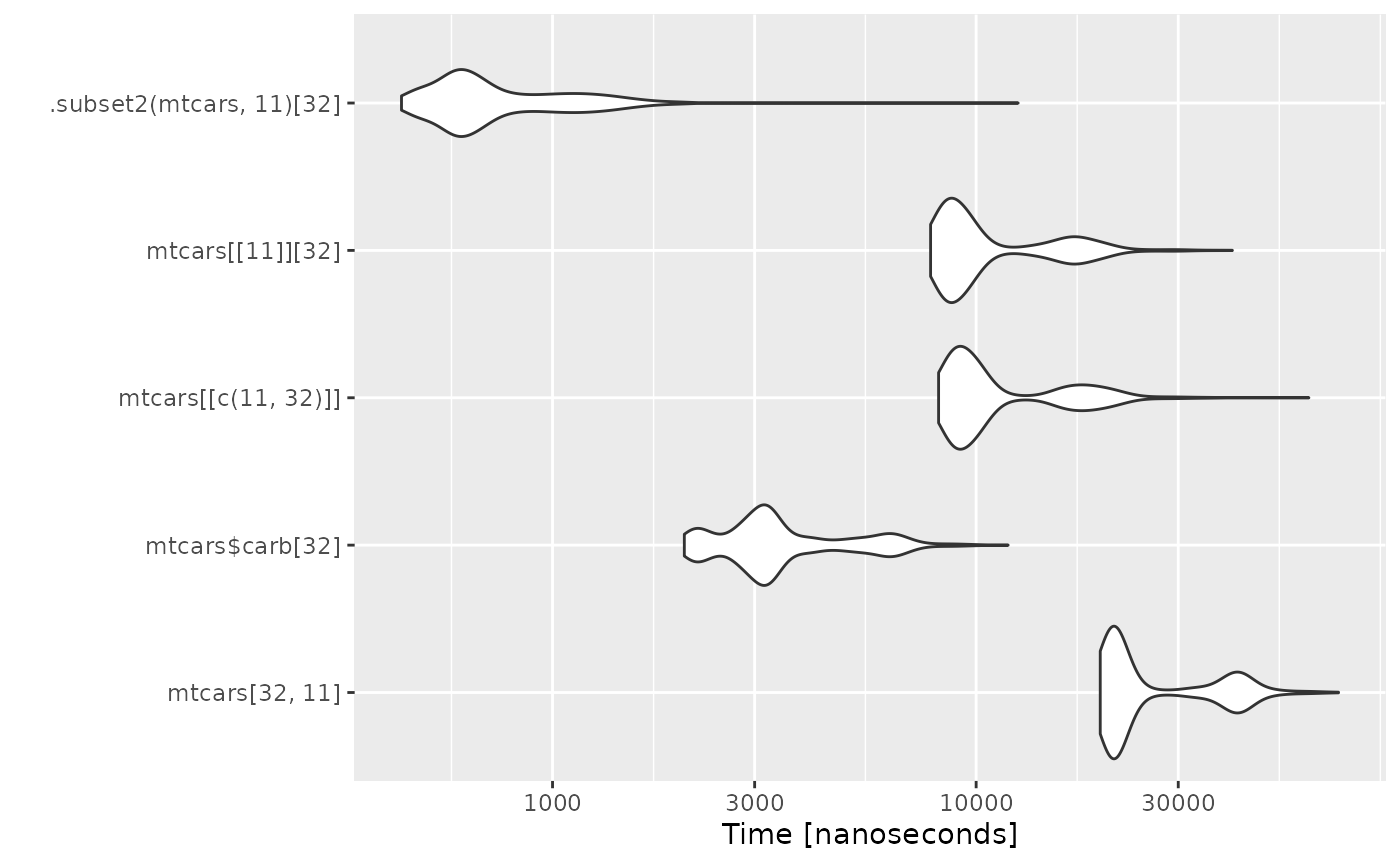
Efficient Value Extraction
Idea
The idea would be to replace the different one-value extraction alternatives by the much faster .subset2 call alternative.
Benchmark
autoplot(microbenchmark(
mtcars[32, 11],
mtcars$carb[32],
mtcars[[c(11, 32)]],
mtcars[[11]][32],
.subset2(mtcars, 11)[32],
times = 1000L
))
Drawback
For some R classes, the
[[ ]]operator and.subsetwork differently. For instance, they seem to be equivalent fordata.framebut are not the same formatrixclass.Moreover, both
[[ ]]and.subset2are functions and in R, any function can be overwritten. Thus the above optimization can be made to fail just by redefining, say, the.subset2function.